 Case Study 3: A group-based analysis of the C. elegans connectome¶
Case Study 3: A group-based analysis of the C. elegans connectome¶
Pyntacle version: 1.0
Table of Contents¶
- Introduction
- Data description and network loading
- The connectome as a rich club of neurons
- Evaluate the group centrality of the rich club core
- Group search of alternative groups of nodes that maximize group degree
- Explore the hierarchy of nested groups inside the rich club using group degree
- Conclusions
- Appendix - Save the binary final graph for later use
Note to the reader: the purpose of these case studies is not scientific, but demonstrative of the functionalities of Pyntacle.
Introduction ¶
This case study revolves around the network-based analysis of the first (and most detailed) wiring diagram (also known as connectome) of the neuronal connections of a fairly complex multicellular organism, the small nematode Caenorhabditis elegans. This systems has been widely studied, even resorting to Graph Theory, to identify emergent properties of neurons (see, for example, the articles by Towlson et al. and by Bentley et al.). Topological analysis of the connections among neurons contributed to understand the spatial architecture and many biological features of this neuronal system, and thus to hypothesize that these and other evidences might be applied also to more complex neuronal systems, such as those of mammalians. This topic is today an open debate. Albeit been widely characterized, there is a gap in the understanding of the role that groups of neurons (nodes, in the connectome) play within the whole network. Topological metrics tailored to the assessment of the importance of groups of nodes, such as the one proposed by Borgatti and Everett, could cover this gap.
This case study takes inspiration from the study of Towlson et al., that identified a rich club in the connectome as the group of top-ranked neurons in terms of their local centrality. We will make use of the Pyntacle's Python library to first replicate the findings of Towlson and, then, to identify other topologically important groups of nodes, possibly including the rich club's members.
In brief, we will:
- Load the connectome into a Python interactive environment.
- Compare our connectome with the one published here.
- Study the rich club of neurons by means of group-based centrality indices.
- Explore the possibility that other groups of neurons smaller or than, as big as, the rich club exist that exhibit higher group centrality values.
- Search for important subgroups of the rich club.
And finally:
- Save the connectome to a graph object for later use.
All the data of this case study are available as an archive here, along with a Python script that contains all commands necessary to repeat this analysis outside of this notebook.
We recommend the reader to have a look at our short introduction on group-centrality indices and to our Pyntacle Quick Start guide before reading this case study, to get acquainted with the metrics that will be used here and with some of the basic Pyntacle library functionalities.
Data description and network loading ¶
We will make use of the neuronal wiring diagram of the hermaphrodite C. elegans that was firstly described in the paper by Varshney et al.. It was downloaded from the WormAtlas database on July 2018. (http://www.wormatlas.org/neuronalwiring.html, section 2.1)
The neuronal wiring file consists of pairs of neurons per line, the synapse type (i.e., sending, receiver), and other ancillary information. The original connection map is made of 283 neurons and 6415 connections. We turned them to a demultiplexed graph by first collapsing multiple connections among neruons into one link (disregarding of their direction). Moreover, we discarded all links representing the neuro-muscular junctions (NMJ) or involving the node named NMJ, as it is a placeholder for the final muscular stimuli, and retained only the links classified as sending (S, Sp) and receivers (R, Rp). The resulting network is available here as a SIF file (see the file formats guide for a full description of the SIF format). The final graph consists of 279 nodes, 1960 edges and has a single component.
Assuming that the Python console (or an interactive notebook) is in the same directory of the connectome file, we can easily import it into Pyntacle as an igraph.Graph object with the following command:
from pyntacle.io_stream.importer import PyntacleImporter as pyimp
graph = pyimp.Sif("CAEEL_Connectome.sif")
The graph summary shows the correct number of nodes and edges, plus a series of attributes, described in our minimum requirements page, which are initialized when importing the graph.
graph.summary()
The connectome as a rich club of neurons ¶
The graph of the worm connectome is similar to a Small-world network, with a high number of connections between neighboring neurons and short distances among all neurons. These properties can be checked by means of Octopus: the interface between igraph and Pyntacle. Octopus aims at relieving the user from the burden of computing topological indices and assigning their values to the Graph object or to the nodes and edges manually.
By means of Octopus, we will first check the average node degree and the average shortest path lengths of the connectome:
from tools.octopus import Octopus
Octopus.average_degree(graph)
Octopus.average_global_shortest_path_length(graph)
The results of these methods are automatically labeled as average_degree and average_global_shortest_path_length, respectively, and added to the graph as graph attributes.
Note that average_global_shortest_path_length differs from average_shortest_path_length as it averages all the geodesics among any node pairs in the graph, while the latter computes the average distance from a specific node to the other nodes of a graph.
From their values:
graph["average_degree"]
graph["average_global_shortest_path_length"]
we can notice high degree, in the average, and short paths between nodes. As anticipated, these are peculiar characteristics of small-world systems.
This connectome graph (as other real complex systems, see Colizza et al) is partitioned into two guilds: a rich club of nodes, which is an elite clique of high-degree hubs, and a large poor periphery.
In their paper, Towlson et al. identified a rich club within the C. elegans connectome made by 287 neurons and 2287 synaptic connections. It is worth noticing that this connectome has 352 more connections than our connectome model, since we removed all the connections representing the electric junctions (EJ). We will explore the impact of this choice later in this chapter.
The authors identified the rich club by normalizing the degree of the top-ranked nodes in the network over the degree of the top-ranked nodes in a 1000 random networks with the same number of nodes and edges than the connectome, thereby identifying the rich club of neurons as the one with at least one standard deviation (σ) over the rich club coefficient (we refer the reader to the original paper for more details on this). In the end, the authors identified a group of 14 high-degree nodes, made exclusively by interneurons, with a strong deviation from the normalized rich club coefficient, thus choosing neurons whose degree in the network strongly differs from their random counterparts. They are described in the following table:
| Neuron ID | $$\it{k_T}$$ | Rich Club | Function |
|---|---|---|---|
| AVAR | 94 | 3σ | Head interneuron; role in locomotor decisions |
| AVAL | 93 | 3σ | Head interneuron; role in locomotor decisions |
| AVBL | 76 | 3σ | Head interneuron; role in locomotor decisions |
| AVBR | 75 | 3σ | Head interneuron; role in locomotor decisions |
| AVER | 57 | 3σ | Head interneuron; role in locomotor decisions |
| AVDR | 56 | 3σ | Head interneuron; role in locomotor decisions |
| AVEL | 56 | 3σ | Head interneuron; role in locomotor decisions |
| PVCL | 55 | 3σ | Head interneuron; role in locomotor decisions |
| PVCR | 53 | 3σ | Head interneuron; role in locomotor decisions |
| DVA | 51 | 3σ | Head interneuron; role in locomotor decisions |
| AVDL | 45 | 3σ | Head interneuron; role in locomotor decisions |
| AIBR | 39 | 2σ | Head interneuron |
| RIBL | 38 | 1σ | Head interneuron |
| RIAR | 37 | 1σ | Head interneuron |
Where $\it{k_T}$ is the degree of the neurons evaluated by Towlson et al. and the Rich Club column reports the distance, expressed in standard deviations from their random counterpart. This allows to identify a core of 11 neurons that are at at least 3 times the standard deviation (3σ) from the normalized rich club coefficient of the graph. This core is depicted in red when plotting the connectome with an external tool (Cytoscape), while the neurons distant 2σ and 1σ are depicted in orange and blue, respectively:
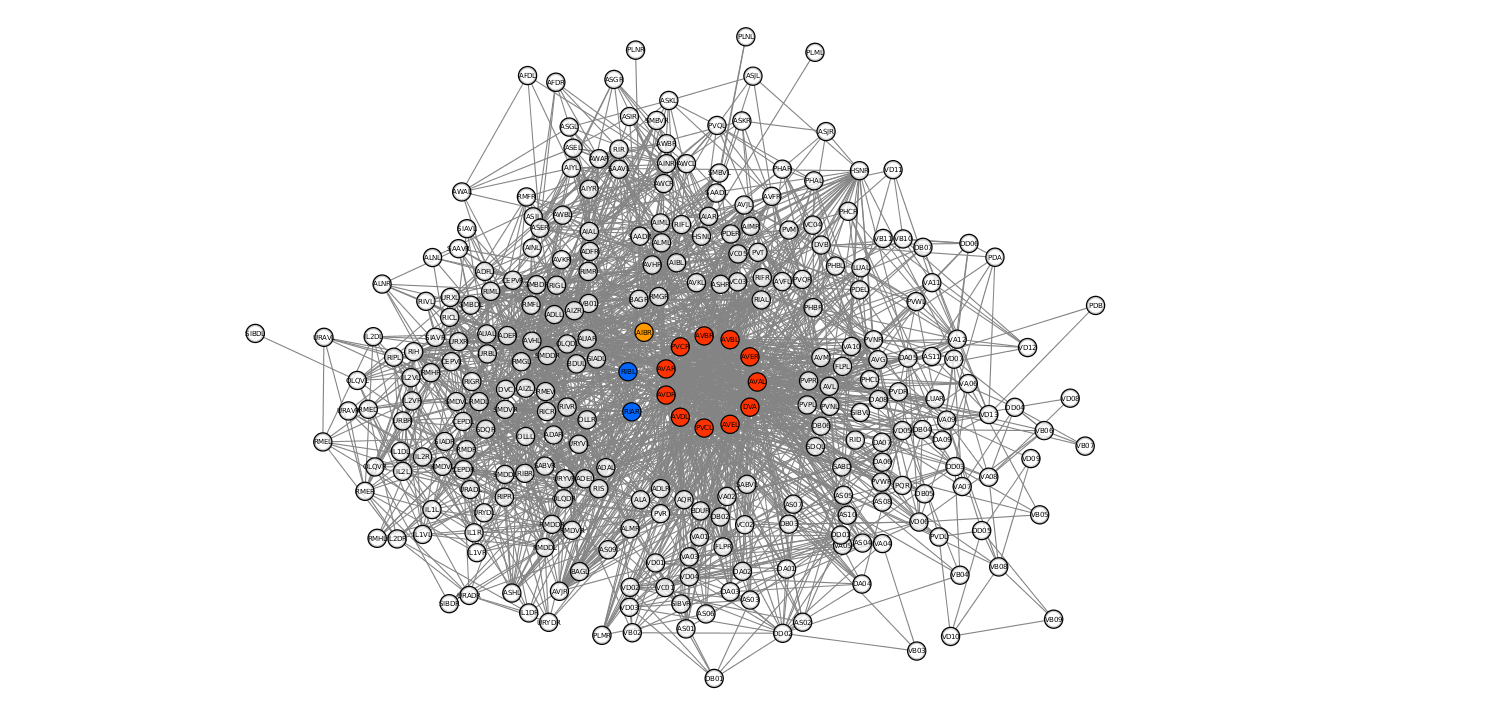
As mentioned before, the absence of edges that represent the electric junctions in our dataset compared to the connectome model built by Towlson is remarkable. This difference may actually lead to different conclusions. One of these is that some of the rich club members may be deprived of meaningful connections that can be, in turn, detrimental in terms of topological centrality.
We can evaluate this difference by sorting by degree the nodes of our connectome and of the Towlson's connectome that belong to the rich club. We do this by means of the degree method of Octopus:
Octopus.degree(graph)
and then combine results into a pandas DataFrame:
import pandas as pd
df = pd.DataFrame({"Degree":graph.vs["degree"]}, index=graph.vs["name"])
df = df.sort_values(by=["Degree"], ascending=False)
Looking at the top 14 neurons of the DataFrame:
df.head(14)
We notice that the top ranked degree nodes are the same except for RIBL, that is replaced by HSNR (a motor neuron, $k_P$=XX). Moreover, the degree rank slightly differs between the two.
If we compare the degree of nodes of the rich club found by Towlson with that of nodes in our connectome, and measure their differences ($\Delta_k$), we obtain the following table:
| Neuron ID | $$\it{k_T}$$ | $$\it{k_P}$$ | $$\Delta_k$$ |
|---|---|---|---|
| AVAR | 94 | 85 | 8 |
| AVAL | 93 | 83 | 10 |
| AVBL | 76 | 56 | 10 |
| AVBR | 75 | 51 | 14 |
| AVER | 57 | 50 | 7 |
| AVDR | 56 | 53 | 3 |
| AVEL | 56 | 50 | 6 |
| PVCL | 55 | 52 | 3 |
| PVCR | 53 | 49 | 4 |
| DVA | 51 | 48 | 3 |
| AVDL | 45 | 42 | 3 |
| AIBR | 39 | 34 | 5 |
| RIBL | 38 | 26 | 8 |
| RIAR | 37 | 36 | 1 |
We can conclude that, despite the most connections loss regards the rich club core and not the other 3 members (AIBR, RIBL, RIAR), the small connection loss in our connectome model leads to as different ranking of the nodes in the connectome. Nevertheless, the top-ranked degree nodes are still the most informative neurones, the one belonging to the 3σ rich club "core". For this reason, we will focus our analysis from here on on this subgroup.
Evaluate the group centrality of the rich club core ¶
While the conditions for defining a rich club are well-established, little is knwon on the centrality that the members of a rich club should hold in a network. This is crucial for assessing the overall importance of the rich club in the connectome and to further characterize its relevance.
We can try to fill this gap here by measuring the group centrality of the rich club'members using the 3 group centrality indices implemented in Pyntacle, namely the group degree, the group closeness and the group betweenness. These are modified version of corresponding local centrality indices. The first measures the overall number of connections that a group has with respet to the rest of the graph, while the other 2 assess the centrality of a group by elaborating on its gedesics distance to the rest of the graph. They all range from 0 to 1, with values closer to 1 meaning a strong centrality of the group. To this aim, we will calculate the group_degree, group_closeness and group_betwenness as implemented in Octopus.
We can first evaluate the group degree of the rich club core by typing:
#list that contains the names of the rich club nodes
rc = ["AVAR", "AVAL", "AVBL",
"AVBR", "AVER", "AVDR",
"AVEL", "PVCL", "PVCR",
"DVA", "AVDL"]
Octopus.group_degree(graph, rc)
#print the group degree attribute along with the node names
graph["group_degree_info"]
Note that the name of the attribute that stores the group degree value of a predefined set of nodes contains the info suffix. This suffix will not be present in case the group degree will be calculated by one of the optimization search algorithms implemented in Pyntacle, since the set of nodes will not be predefined, but contextually found by the algorithms.
Note also that all the metrics of groups will be stored as graph's attributes and thus as Python dictionaries, whose keys are tuples containing the node set, and their matching values being floats representing their group centrality values.
The normalized group degree coefficient of the rich club is 0.63433. Similarly, we can compute the group betwenness of the rich club:
Octopus.group_betweenness(graph, rc)
graph["group_betweenness_info"]
And its group closeness:
Octopus.group_closeness(graph, rc)
graph["group_closeness_minimum_info"]
Note that the group closeness graph attribute name contains a further suffix, minimum. Since the group closeness is calculated as the sum of the inverse distances between a nodes set and the rest of the graph, you may be interested to the (default) minimum, maximum or average distance between the nodes set and the rest of the network. In our connectome, we have considered the minimum distances, as we are interested in the shortest possible communications between the group and the other neurons.
Assembling all the three group centralities into a pandas pd.DataFrame object (downloadable as a tab-delimited file here):
rc_ind = " ".join(rc)
rc_key = tuple(rc)
dfdict = {"group degree": graph["group_degree_info"][rc_key],
"group betweenness": graph["group_betweenness_info"][rc_key],
"group closeness (min)": graph["group_closeness_minimum_info"][rc_key]}
df = pd.DataFrame(dfdict, index=[rc_ind])
df
We noted that, as a group, the rich club is central in terms of the overall distances between it and the rest of the graph, as the group exhibit considerably high values of group degree and closeness. Nevertheless, the group betwenness of the rich club is significantly low (0.27), meaning that, on overall, there is a low number of shortest paths that pass though the rich club core with respect to the rest of the nodes in the graph. This is expected, as we symmetrized the connections by eliminating the directionality of the edges.
The group centrality values that we computed here are of course relative to the rich club. Hence, this does not exclude the presence of other groups, which are not strictly rich clubs, but that exhibit higher centrality values. In the next paragraph, we are going to search for other groups of nodes of the same size as the rich club that maximize the group degree of the connectome.
Group search of alternative groups of nodes that maximize group degree ¶
We have anticipated that there may be other sets of nodes that, together, may exhibit even higher values of group degree. We can search for these groups using the greedy optimization search algorithm implemented in Pyntacle or directly using the GO_group_degree method (it will take some minute on a standard desktop) of Octopus. It will add a group_degree_greedy attribute to the graph.
Note that this process will not necessarily return the best set (or sets) of nodes, as this algorithm is greedy and thus will not explore all the space of solutions. To do that, the user may want to call the BF_group_degree method instead, that launches a parallel, brute-force search. As a note of caution, please consider that a brute-force search may take longer than expected to finish its computation, especially when dealing with sets bigger than 5 nodes.
seed = 1
Octopus.GO_group_degree(graph, 11, seed=seed)
graph["group_degree_greedy"]
We can notice that there is at least one set with a higher group degree (0.82836) than that of the rich club. This table reports the neurons, along with their functions and their degree ($k_P)$ values in the connectome:
| Neuron ID | $k_P$ | Function |
|---|---|---|
| AIBL | 32 | Interneuron |
| AVAR | 85 | Interneuron |
| AVHR | 19 | Interneuron |
| CEPVR | 48 | Sensory |
| HSNL | 24 | Motor neuron |
| PVCL | 52 | Ventral cord interneuron |
| RIAL | 33 | Sensory neuron |
| RIH | 31 | Interneuron |
| RMDR | 21 | Motor neuron |
| RMEL | 9 | Motor neuron |
| VC03 | 22 | Motor neuron |
Some of these neurons are part of the rich club (i.e., AVAR, PVCL), while other "low-degree" nodes are spread across the network. However, these together contribute to obtaine a higher group degree than that of the rich club. Some of the neurons are not interneurons, rather they belong to the motor or sensory classes of neurons, implying that they can reach easily the network periphery. These nodes are highlighted in green in the graphic representation of our connectome model:
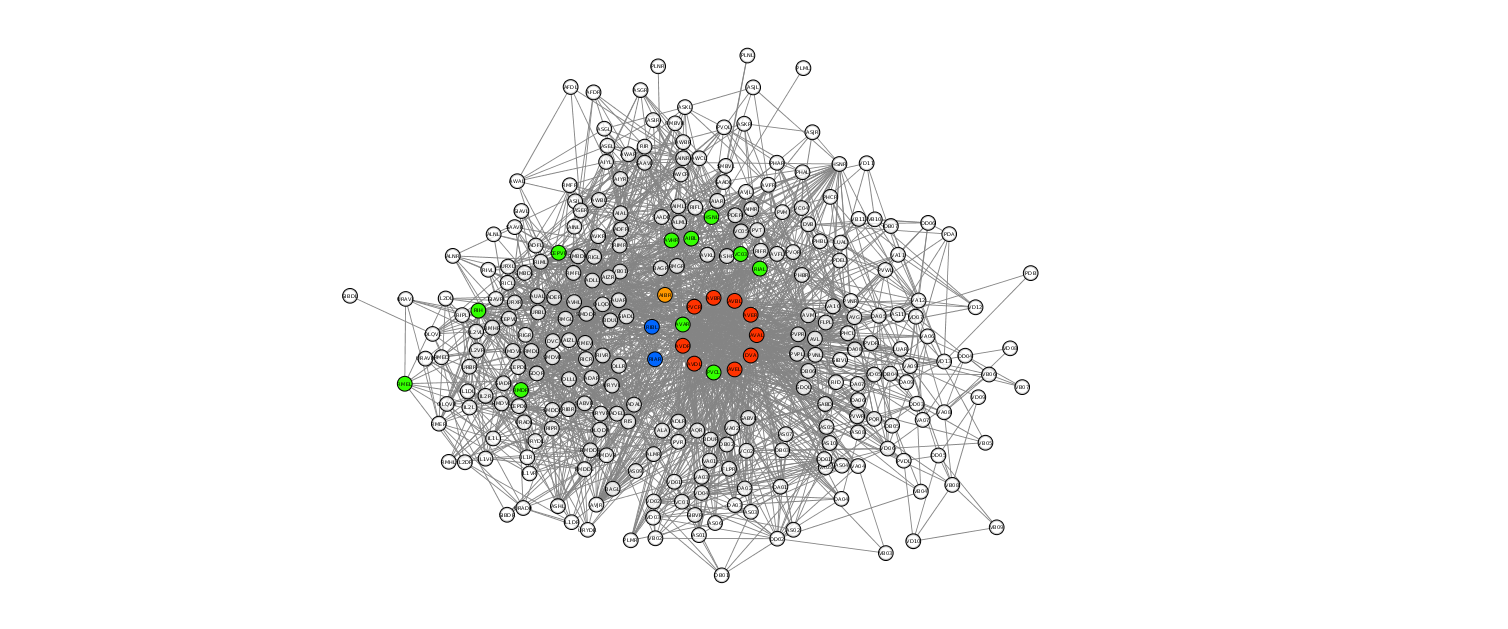
With this, we can partially explain why the group degree of the rich club may not be the highest. The members of the rich club are well connected, if compared with other ndoes of the network. This elevate connectance among members is for sure optimal in terms of neural information exchange and robustness to external insults, but penalizes the rich club when considering it as a group.
This very preliminary study shows that computing the group degree can add additional layers of information about the struture of neuronal networks.
Explore the hierarchy of nested groups inside the rich club using group degree ¶
In the last part of this case study, we focus on another question concerning the rich club and its group degree: does each node in the rich club contribute equally to the group degree of the rich club? In other terms, is there any nested subset of nodes of size $k$ that exhibits a higher group degree value than the group degree values calculated for all the other groups of neurons of same size?
Only for demonstrative purposes, we will calculate the group degree values of all sets of 3 neurons ($k=3$):
import itertools
rc_shuffled = [list(x) for x in itertools.combinations(rc, 3)]
for elem in rc_shuffled:
Octopus.group_degree(graph, elem)
graph["group_degree_info"]
We can then store results in a pandas DataFrame and sort the group degree values of each set in descending order:
gd = graph["group_degree_info"]
gd = gd.values()
df = pd.DataFrame({"node set":["_".join(x) for x in graph["group_degree_info"].keys()], "group degree": list(gd)})
df = df.sort_values(by="group degree", ascending=False)
df
(The dataframe is available as tab delimited file here)
At this point, we can plot the distribution of the group degree values of each set $h$, and compute mean and standard deviation values:
#import all the libraries for plotting
%matplotlib inline
import seaborn as sns
import matplotlib.pyplot as plt
from scipy import stats
import numpy as np
#load the dataset
df = pd.read_csv("CS3_inner_groupdegree_RC.tsv", sep="\t", index_col=0)
df.head()
%matplotlib inline
plt.figure(figsize=(15,8))
sns.set_style('white')
sns.set_context("notebook", font_scale=1.5, rc={"lines.linewidth": 2.5})
ax = sns.distplot(df["group degree"], rug=False, color="green")
ax2 = ax.twinx()
sns.boxplot(x=df["group degree"], ax=ax2, color="darkgreen")
ax2.set(ylim=(-0.5, 10), yticks = [])
sns.despine()
plt.xlim(0,1)
#compute the row mean and stdev
mean = df["group degree"].mean()
stdev = df["group degree"].std()
mean, stdev
We can see here that the distribution of the group degree values for all sets of neurons of size 3 is quite homogeneous around its mean, 0.4, with a standard deviation of 0.04. If we look at the sets $h$ that are distant from this distribution and e xhibhit higher values of group degree, we notice that they always contains at least one among these 3 neurons: AVAR, AVAL and AVBR, the ones with the highest degree scores in ur connectome model. This may suggest that the group degree of these subsets is led by these nodes.
Conclusions ¶
In this case study, we analyzed the neuronal wiring diagram of the nematode C. elegans, through group centrality indices.
In the first part of the study, we compared our network model of the worm connectome with that made by Towlson et al. in a paper that dates back to 2013. By means of Pyntacle, we compared the degree values of the nodes belonging to the rich club and concluded that its core, which was made of 11 nodes, exhibited significantly high and comparable values of degree in both networks. We thus focused on the rich club core and computed its group centrality values, finding that on overall the rich club exhibits high values of group closeness and of group degree, but a low group betweenness. These findings remark that:
- Hierarchically, the rich club is well-placed in the connectome, with a high group closeness value;
- The symmetrization of the network penalizes the rich club in terms of group betweenness, since the majoriy of the shortest paths does not pass through it. Future versions of Pyntacle, which will account for directed and signed graphs, will be able to compute the group betweenness more precisely.
We then tried to find other interesting sets of neurons with the same size of that of the rich club through a greedily-optimized search. Surprisingly, we found that at least one set, made of (almost totally) disconnected nodes, yielded a much higher group degree than that of the rich club. This is mainly due to the fact the the rich club is well-connected among its members, although it is not so well-connected with the rest of the network with respect to other sets of the same size.
Finally, we investigated whether the group degree of the rich club is due to the equal contributions of all its components or of a subset of it. We computed the group degree of all sets of size 3 (arbitary choice) and showed that the group degree was almost comparable among all sets, with some subgroup exhibiting higher group degree values when including at least one of these 3 neurons: AVAR, AVAL and AVBR.
from pyntacle.io_stream.exporter import PyntacleExporter as pyexp
pyexp.Binary(graph, "CS3_binary.graph")
The file can be downloaded here
This concludes our case study 3. If you want to leave a feedback, please contact us.